Section: New Results
Medical Image Analysis
Segmentation of cardiac images from magnetic resonance
Participants : Jan Margeta [Correspondant] , Kristin Mcleod, Antonio Criminisi [MSRC] , Nicholas Ayache.
This work has been partly supported by Microsoft Research through its PhD Scholarship Programme and the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Cardiac imaging, Magnetic resonance, Image segmentation, Maching learning
-
We contributed our previous method to build left ventricle myocardium segmentation consensus based on the STAPLE algorithm [26]
-
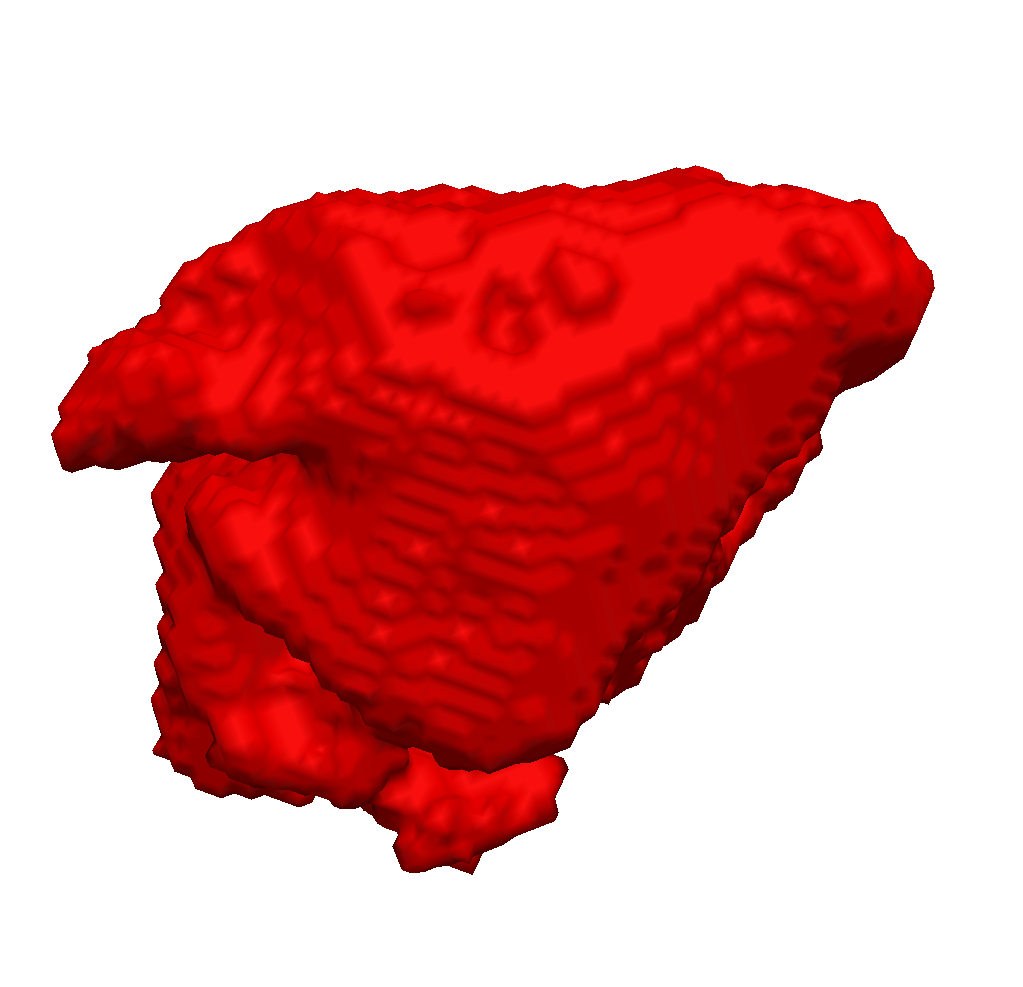
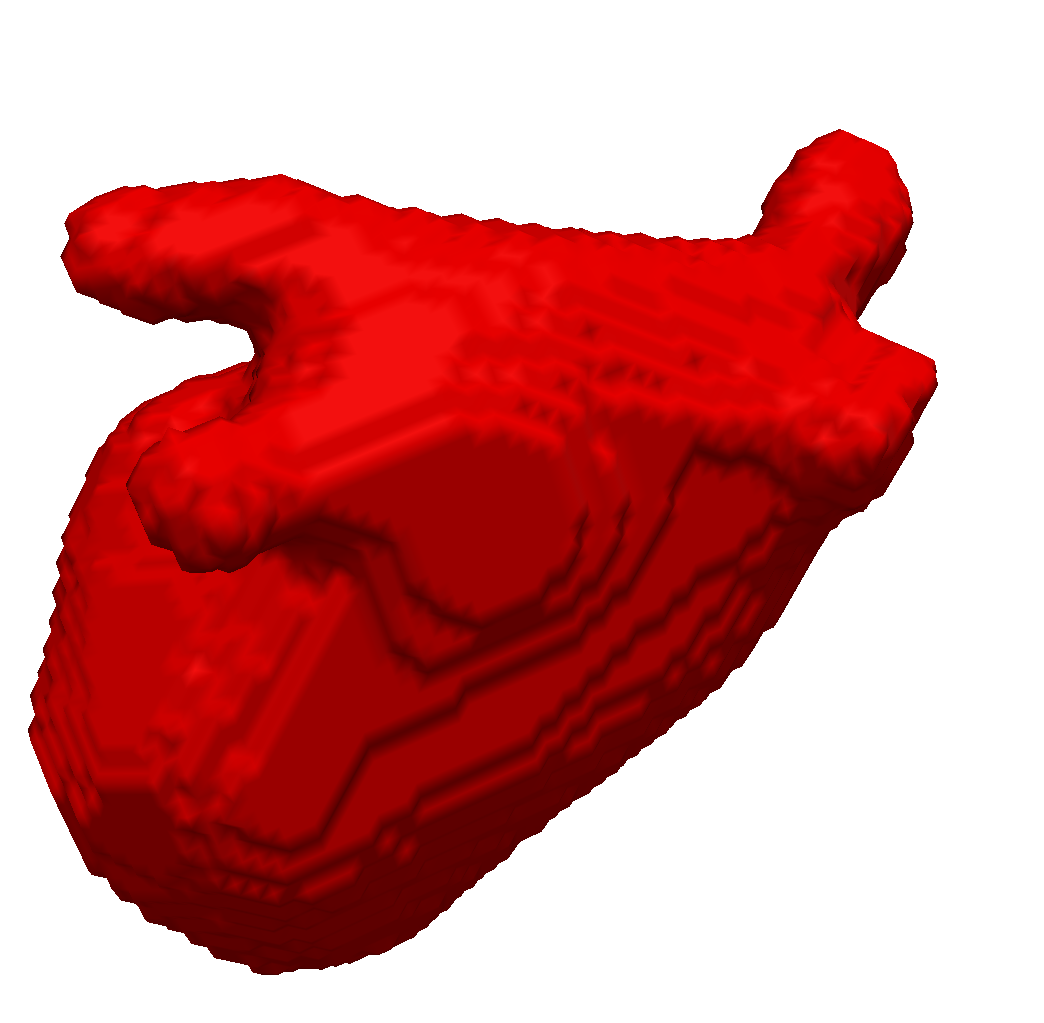
We enhanced our segmentation method with extra features based on the distance transform and image vesselness measures in order to segment left atria (see Fig. 1 ) from 3d MRI images [49] . We participated with this method in the left atrium segmentation challenge at MICCAI 2013.
Brain tumor image processing and modeling
Participants : Bjoern Menze [Correspondant] , Hervé Delingette, Nicholas Ayache, Nicolas Cordier, Erin Stretton, Jan Unkelbach.
We developed a new non-parametric lesion growth model for the analysis of longitudinal image sequences [59] , evaluated the parametric tumor growth model of Konukoglu on longitudinal data, focusing on the relevance of DTI [40] , [57] , and addressed the question of how to detect tumor growth from longitudinal sequences of patients treated with angiogenesis inhibitors using registration techniques [47] . We also completed work for the 2012 MICCAI Challenge on Brain Tumor Image Segmentation (MICCAI-BRATS 2012) [79] , where we also tested some of our own brain tumor image segmentation models based on random forests [42] and patch regression [38] , we also participated in MICCAI-BRATS 2013 in Nagoya, Japan [67] .
Further developing the random forest framework for medical computer vision tasks
Participants : Bjoern Menze [Correspondant] , Matthias Schneider, Ezequiel Geremia, Rene Donner, Georg Langs, Gabor Szekely.
Methodological contributions include the further development of the random forest framework. We introduced the “spatially adaptive” random forest (SARF) classifier [42] , and evaluated Hough regression forests for interest point detection in whole body CT image analysis, as well as for vessel detection and tracking [54] . We also evaluated alternative patch-based methods for whole body image registration [41] . As a related community effort, we organized the MICCAI-MCV workshop, also in conjunction with the MICCAI conference in Nagoya, Japan [65] .
Statistical Analysis of Diffusion Tensor Images of the Brain
Participants : Vikash Gupta [Correspondent] , Nicholas Ayache, Xavier Pennec.
Diffusion Tensor Imaging of the Brain, Tractography, Super-resolution, Statistical analysis
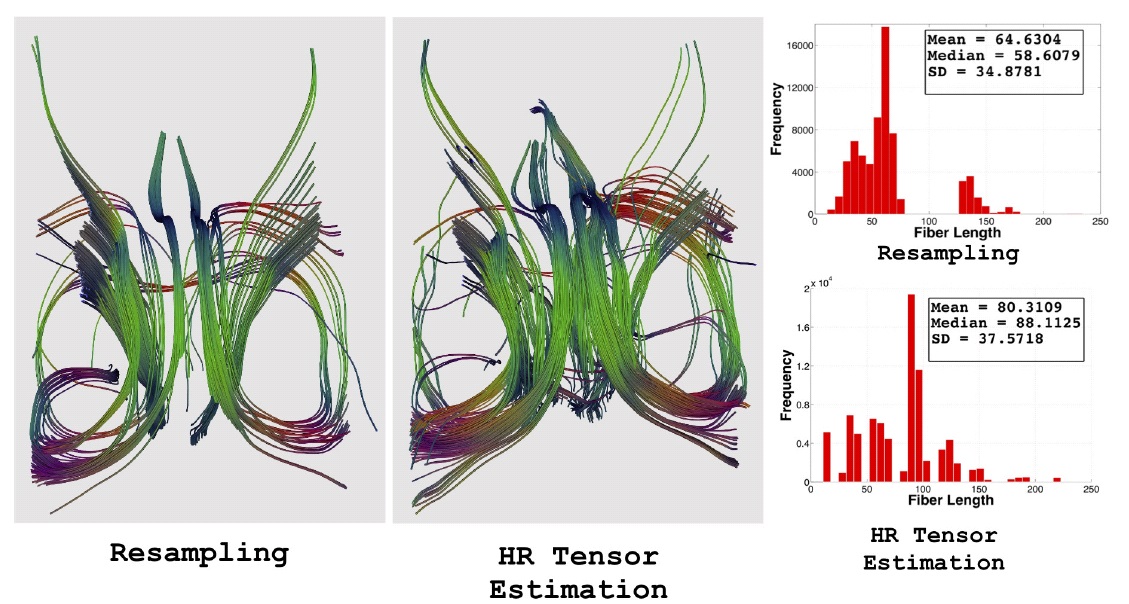
Diffusion tensor imaging (DTI) is gaining interest as a clinical tool for studying a number of brain diseases pertaining to white matter tracts and also as an aid in neuro-surgical planning. Unfortunately, in a clinical environment, diffusion imaging is hampered by the long acquisition times, low signal to noise ratio and a prominent partial volume effect due to thick slices. We are developing a framework for increasing the resolution of the low-resolution clinical CTI images. The method uses a maximum likelihood strategy to account for the noise and an anisotropic regularization prior to promote smoothness in homogeneous areas while respecting edges. The technique is called Higher Resolution Tensor Estimation and it uses a single clinical acquisition to produce high resolution tensor images. We aim to replace resampling techniques used for tensor normalization in population based studies, with the present method. The method itself along with quantitative results on tractography 2 were presented in MICCAI 2013 [45] .
|
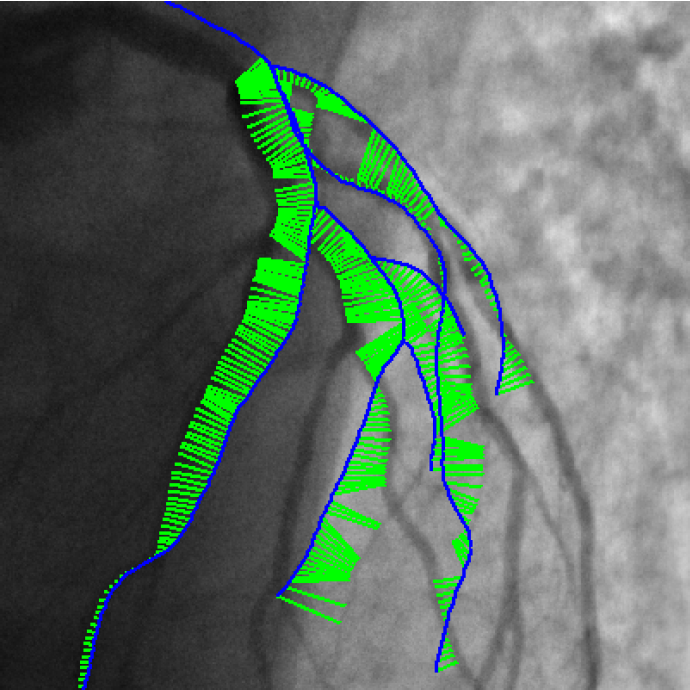
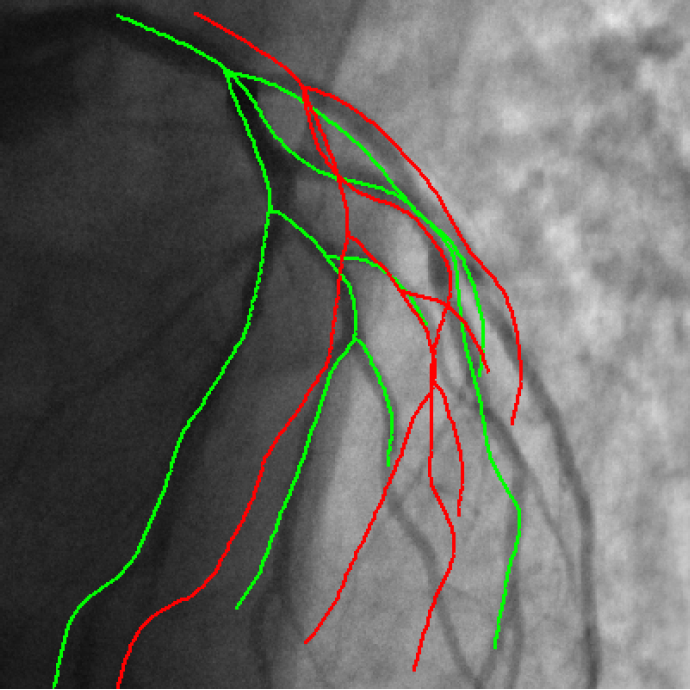
3D/2D coronary arteries registration
Participants : Thomas Benseghir [Correspondant] , Grégoire Malandain, Régis Vaillant [GE-Healthcare] , Nicholas Ayache.
This work is done in collaboration with GE-Healthcare (Buc).
3D/2D registration; computed tomography angiography; CTA; X-ray fluoroscopy; coronary arteries
Endovascular treatment of coronary arteries involves catheter navigation through patient vasculature. Projective angiography guidance is limited in the case of chronic total occlusion where the occluded vessel can not be seen. Integrating standard preoperative CT angiography information with live fluoroscopic images addresses this limitation but requires alignment of both modalities.
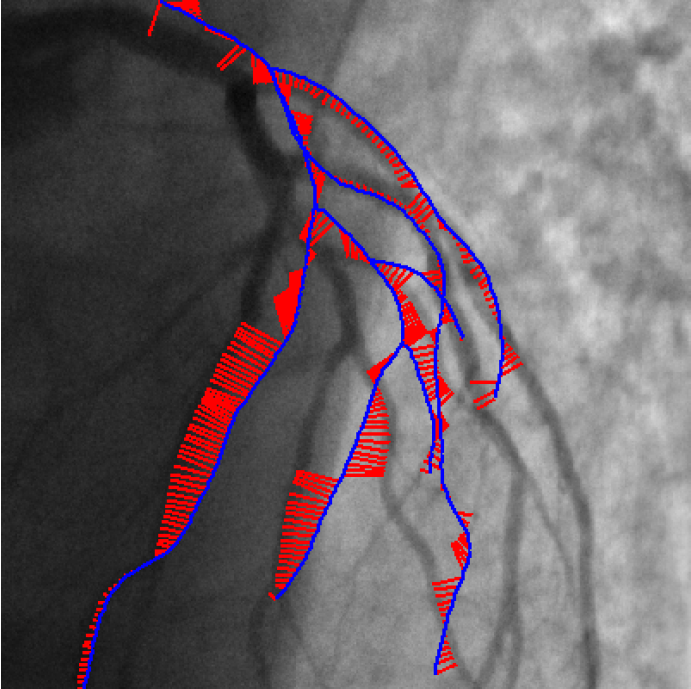
We published the Iterative Closest Curve (ICC) algorithm [36] in the MICCAI 2013 conference :
-
The ICC-algorithm mimics the ICP-algorithm (P.J. Besl and N.D. McKay. A method for registration of 3-D shapes), curves being considered instead of points.
-
Contrary to closest point pairing, the resulting pairings assure a topological and geometrical coherence since a curve is paired to another one (cf Figure 3 ).
-
The developed method can deal with differences in both datasets by considering outlier rejection at the level of curve and the level of point.
|
Automatic Registration of Endoscopic Images
Participants : Anant Vemuri [Correspondant] , Stéphane Nicolau [IHU Strasbourg] , Luc Soler [IHU Strasbourg] , Nicholas Ayache.
This work is performed in collaboration with IHU Strasbourg.
Image registration; Endoscopic imaging; Biopsy Relocalization
The screening of cancer lesions in the oesophagus involves obtaining biopsies at different regions along the oesophagus. The localization and tracking of these biopsy sites inter-operatively poses a significant challenge for providing targeted treatments.
Our work [61] introduces a novel framework for accurate re-positioning of the endoscope at previously targeted sites:
-
it includes an electromagnetic tracking system in the loop and provides a framework for utilizing it for re-localization inter-operatively.
-
We have shown on three in-vivo porcine interventions that our system can provide accurate guidance information, which was qualitatively evaluated by five experts.